Mass spectrometry is currently one of the most widely used analytical methods for protein analysis in biomedical research. Proteomics which is large-scale protein analysis by mass spectrometry has become an important method for identification and characterization of medically relevant proteins which can serve as biomarkers for disease onset, progression, and severity. Mass spectrometry protein analysis is now routinely used in all biomedical research areas where it is applied in discovery and validation in cancer biomarker research. This powerful technique can be used to monitor response to treatment, or tailor treatment with the aim of obtaining better results for patients. The ability to analyze proteins using mass spectrometers has positively impacted cancer research including breast, skin, brain, prostate, ovarian, bladder and pancreatic cancers.
Several scientific papers published in 2020 including those by Uzzaman et al [1] and Sun et al [2] describe mass spectrometry protein analysis methods to support cancer research. Also, Liu et al [3] used a combination of MS-intensive methods such as isobaric tags for relative and absolute quantitation with two-dimensional liquid chromatography-tandem mass spectrometry (iTRAQ-2D LC-MS/MS) and 1D-targeted LC-MS/MS, on serum samples from healthy people (normal control, NC), patients with benign diseases (BD), and Pancreatic Cancer (PC) patients to identify novel biomarkers of PC. They identified more than 1000 proteins, verified 142 differentially expressed proteins, and finally targeted four proteins for absolute quantitation in 100 serum samples. The novel biomarker panel of apolipoprotein E (APOE), inter-alpha-trypsin inhibitor heavy chain H3 (ITIH3), apolipoprotein A-I (APOA1), apolipoprotein L1 (APOL1), combining with CA19-9, statistically-significantly improved the sensitivity (95%) and specificity (94.1%), outperforming CA19-9 alone, for the diagnosis of PC. Radu et al [2] reports that mass spectrometry provides a powerful and sensitive approach for the analysis of biological systems, and dedicated instruments are available for different classes of analytes, such as proteins, metabolomes, genomes (DNA), transcriptomes, etc.
ITSI-Biosciences offers comprehensive mass spectrometry protein analysis services to support all cancer research. Specifically, services such as global protein analysis, targeted protein identification, protein quantitation, biomarker discovery, global protein expression profiling and post translational modification (PTM) mapping can be applied to all areas of cancer research. ITSI has been offering such mass spectrometry services since 2005 and has supported over 1000 cancer research projects conducted by scientists in academic, biotechnology and medical research laboratories. Human and non-human samples can be analyzed, and thousands of proteins and candidate biomarkers have been identified by mass spectrometry. As little as 1ug of total protein can be analyzed. There are several complementary technologies including Agilent Bioanalyzer, Luminex xMAP and GE 2D-DIGE platforms which are available at ITSI to support mass spectrometry-based cancer research. Figure 1 is a generalized ITSI LC-MS/MS workflow which has been applied to the study of several cancers [4-9].
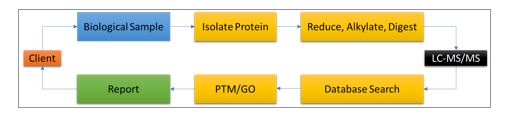
Figure 1: Generalized workflow for biological sample analysis by liquid chromatography – tandem mass spectrometry (LC-MS/MS) at ITSI. Post translational modification mapping (PTM) and gene ontology classification are additional services that add value to the data obtained by mass spectrometry.
ITSI Biosciences uses high performance LC-MS/MS ion trap technologies to offer outstanding mass spectrometry protein analysis services. All biological sample types can be analyzed including 1D-gel slices, 2D gel spots, protein lysate, blood, serum, plasma, saliva, urine, whole tissue, western blot membranes, formalin-fixed paraffin embedded samples and laser micro-dissected samples. The standard mass spectrometry workflow for protein analysis may involve processing samples in a specialized way to isolate proteins prior to mass spectrometry. There could be a cleanup step using our in-house ToPREP total protein precipitation kit to remove all interfering substances. The precipitated samples are resuspended in a digestion buffer and digested with trypsin or other enzyme as needed to achieve the goals of the analysis. The digested peptides are then separated by an inline liquid chromatography step at a nanoliter flow rate, to separate the peptides by reverse phase chromatography on a C18 column prior to an electrospray ionization step. The ionized peptides are then analyzed using a high-performance mass spectrometers including LTQ XL, QExactive orbitrap or Fusion Lumos mass spectrometer. The raw data files are further processed using Proteome Discoverer to identify peptides, proteins, and common peptide modifications, e.g. acetylation, methylation, and phosphorylation. Additionally, Gene Ontology can be performed to define the molecular functions, cellular locations and biological process that the identified proteins may carry out.
The typical turnaround time is 1-2 weeks for all mass spectrometry protein analysis services depending on the project. For more information, contact ITSI-Biosciences via Email; itsi@itsibio.com, Phone; 814-262-7331 or Fax; 814-262-7334.
Reference
- Uzzaman A, Zhang X, Qiao Z, Zhan H, Sohail A, Wahid A, Shang Z, Guan X, Cao CX, Xiao H (2020). Discovery of small extracellular vesicle proteins from human serum for liver cirrhosis and liver cancer. 2020 Aug 21:S0300-9084(20)30198-X. doi: 10.1016/j.biochi.2020.08.013. Online ahead of print.PMID: 32835735
- Sun S, Zhang H, Wang Y, Gao J, Zhou S, Li Y, Han S, Li X, Li J (2020). Proteomic Analysis of Human Esophageal CancerUsing Tandem Mass Tag Quantifications. Biomed Res Int. 2020 Aug 7;2020:5849323. doi: 10.1155/2020/5849323. eCollection 2020.PMID: 32832552
- Liu X, Zheng W, Wang W, Shen H, et al (2017). A new panel of pancreatic cancer biomarkers discovered using a mass spectrometry-based pipeline. Br J Cancer. 2017 Dec 5; 117(12): 1846–1854. doi: 1038/bjc.2017.365.
- Radu Albulescu, Andrei Jose Petrescu, Mirela Sarbu, et al (2019). Mass Spectrometry for Cancer Biomarkers, Proteomics Technologies and Applications, Ibrokhim Y. Abdurakhmonov, IntechOpen, DOI: 10.5772/intechopen.85609. Available from: https://www.intechopen.com/books/proteomics-technologies-and-applications/mass-spectrometry-for-cancer-biomarkers
- Somiari RI, Sullivan, A, Russell, S, Somiari, S, Hu, H, Jordan, R, George, A, Katenhusen, R, Buchowiecka, A, Arciero, C, Brzeski, H, Hooke, J, Shriver, C. (2003). High throughput proteomic analysis of infiltrating ductal carcinoma of the breast. Journal of Proteomics. 10 (3): 1863 – 1873
- Somiari, RI, Somiari SB, Russell S and Shriver CD. (2005) Proteomics of breast carcinoma. J. Chromatography B, Analyt Technol Biomed Life Sci. 815: 215-225.
- Boyiri T, Somiari RI, Russell, S, Aliaga, C and El-Bayoumy K (2009). Proteomics of rat prostate lobes treated with 2-N-hydroxylamino-1-methyl-6-phenylimidazo [4,5-b] pyridine and 5α-dihydrotestosterone. International Journal of Oncology 35: 559-567.
- Bortner J; Das A; Umstead T; Freeman W; Somiari R; Aliaga C; Phelps D; El-Bayoumy K (2009). Down-Regulation of 14-3-3 Isoforms and Annexin A5 Proteins in Lung Adenocarcinoma – Induced by the Tobacco-Specific Nitrosamine NNK in the A/J Mouse Revealed by Proteomic Analysis. Journal of Proteome Research 8(8): 4050-4061
- Arciero C, Somiari, S, Shriver C, Brzeski, H, Jordan, R, Hu H, Ellsworth D, Somiari, RI (2003). Functional relationship and gene ontology classification of breast cancer biomarkers. International Journal of Biological Markers 18 (4): 241-272.
